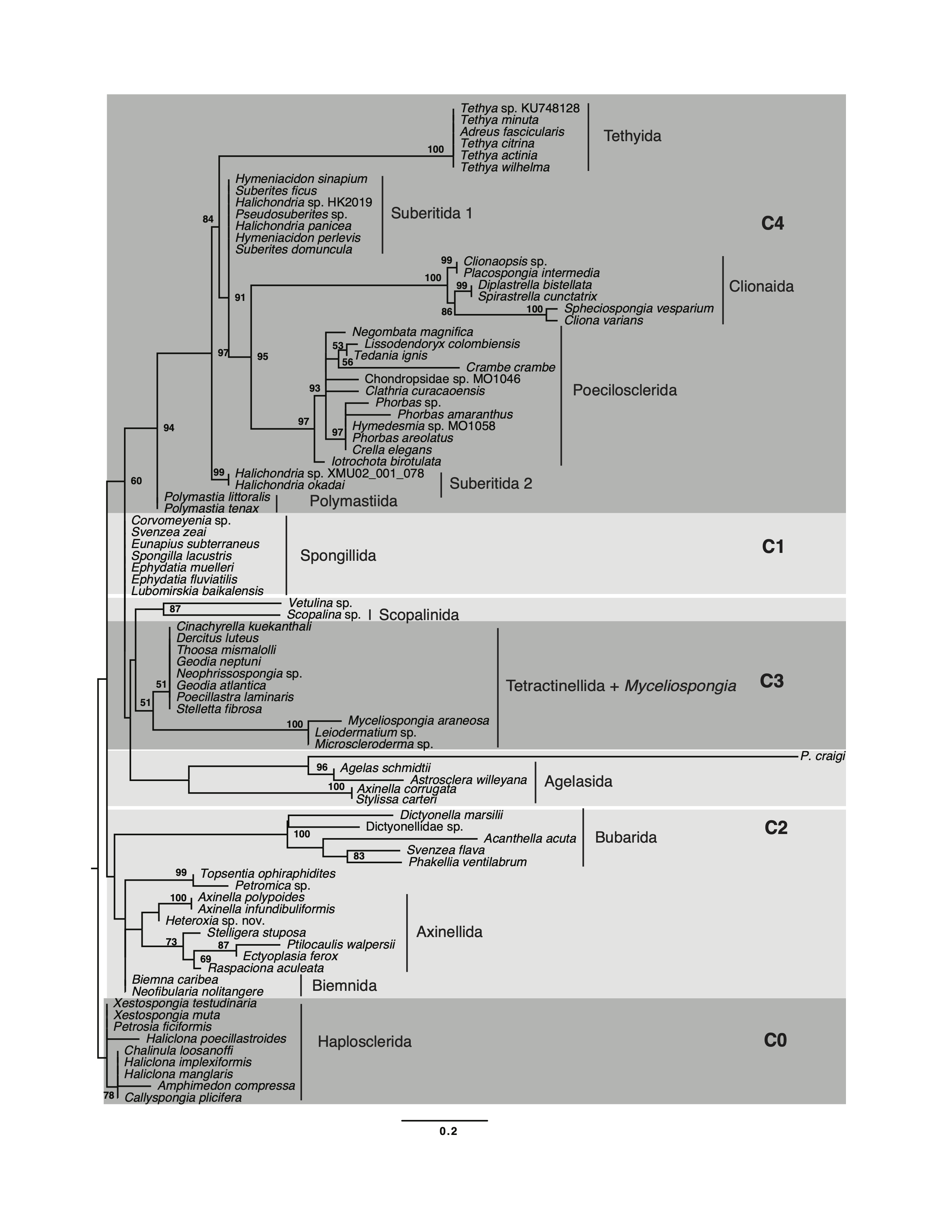
Demosponge phylogeny based on mitochondrial gene orders
Maximum likelihood reconstruction of demosponge relationships based on the multistate encoding of the gene boundary data. Please note that there are many fewer characters in the gene order dataset comparing to the sequence data. Analysis was conducted in RAxML-NG v. 1.1.0 using MULTI13_MK model.
Options
- Download this figure in the PDF format;
- Download the tree in Newick format;
- Open this tree in iTOL;
- Download the alignment used to build the tree.

Content created by Lavrov Lab at Iowa State University.
Hosted by GitHub Pages.
Jekyll theme based on Millidocs.
Except where otherwise noted, content on this site is licensed under a Creative Commons Attribution 4.0 International License.


