Time estimates for major diversification events in demosponge phylogeny
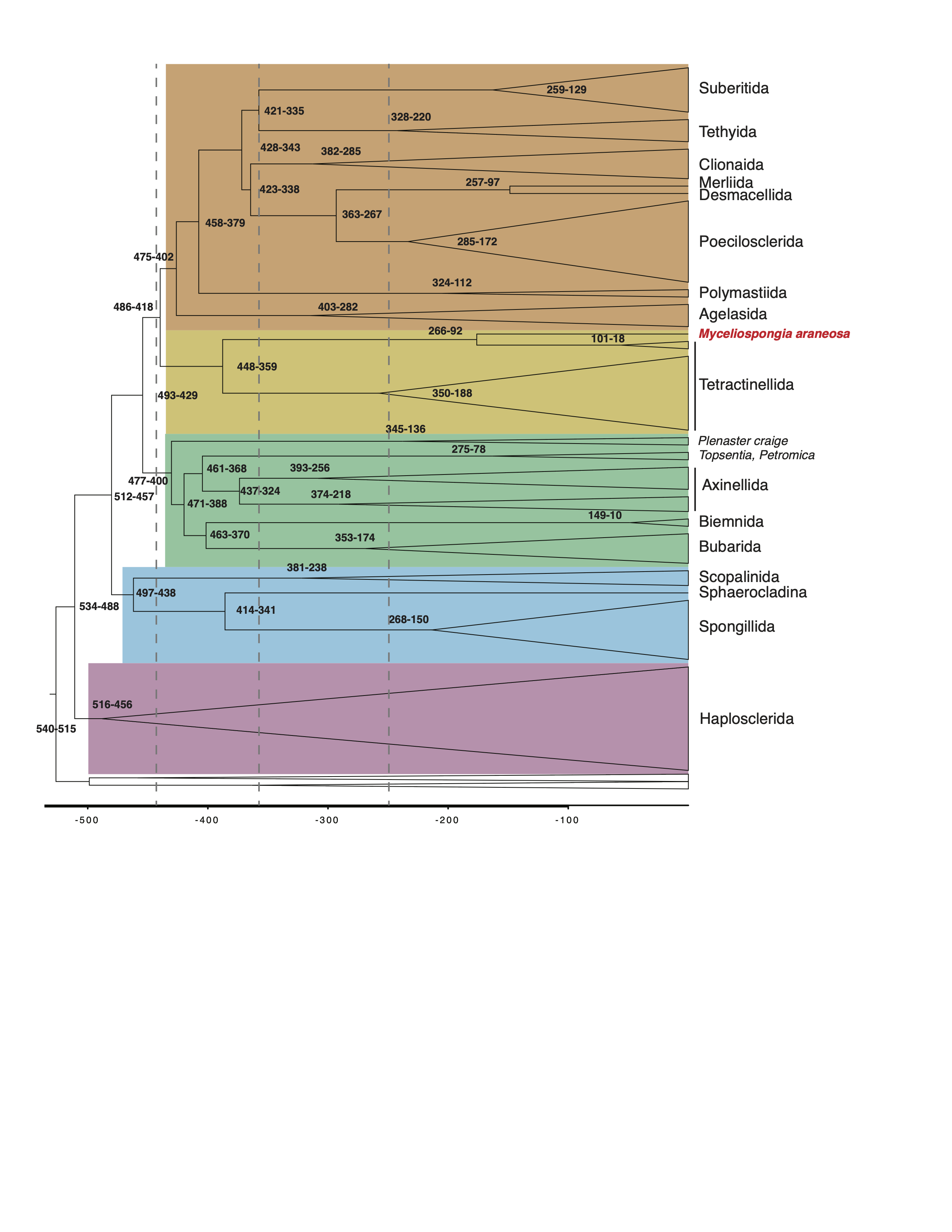
Simplified time calibrated phylogeny with only heterosleromorph orders being labeled.
Time constraints
The following time constraints were used in the analysis:
- a uniform prior was placed on the root of the demosponges between 541 and 515 MY;
- the split between Spongillida and Vetulina sp. was timed at between 410 and 298 MY;
- the origin of the crown group Lake Baikal sponges was constrained between 30 and 6 MY. All calibration ranges were specified as soft bounds, which allocates 0.025 of the total probability outside the specified bounds.
Dataset
The same dataset was utilized for the time analysis as for the phylogenetic analysis.
Phylobayes analysis
PhyloBayes 4.1c was used for the molecular clock analysis on the fixed tree topology inferred from translated mitochondrial coding sequences. Two chains were run for 20,000 cycles and convergence was assessed by estimating discrepancies and effective sizes for continuous variables in the model. Log-normal autocorrelated relaxed clock model, a generalized time-reversible amino acid substitution matrix, a Dirichlet mixture profile, and a discrete gamma distribution with four categories were used.
The full tree can be seen here.

Content created by Lavrov Lab at Iowa State University.
Hosted by GitHub Pages.
Jekyll theme based on Millidocs.


