Publications
Highlights
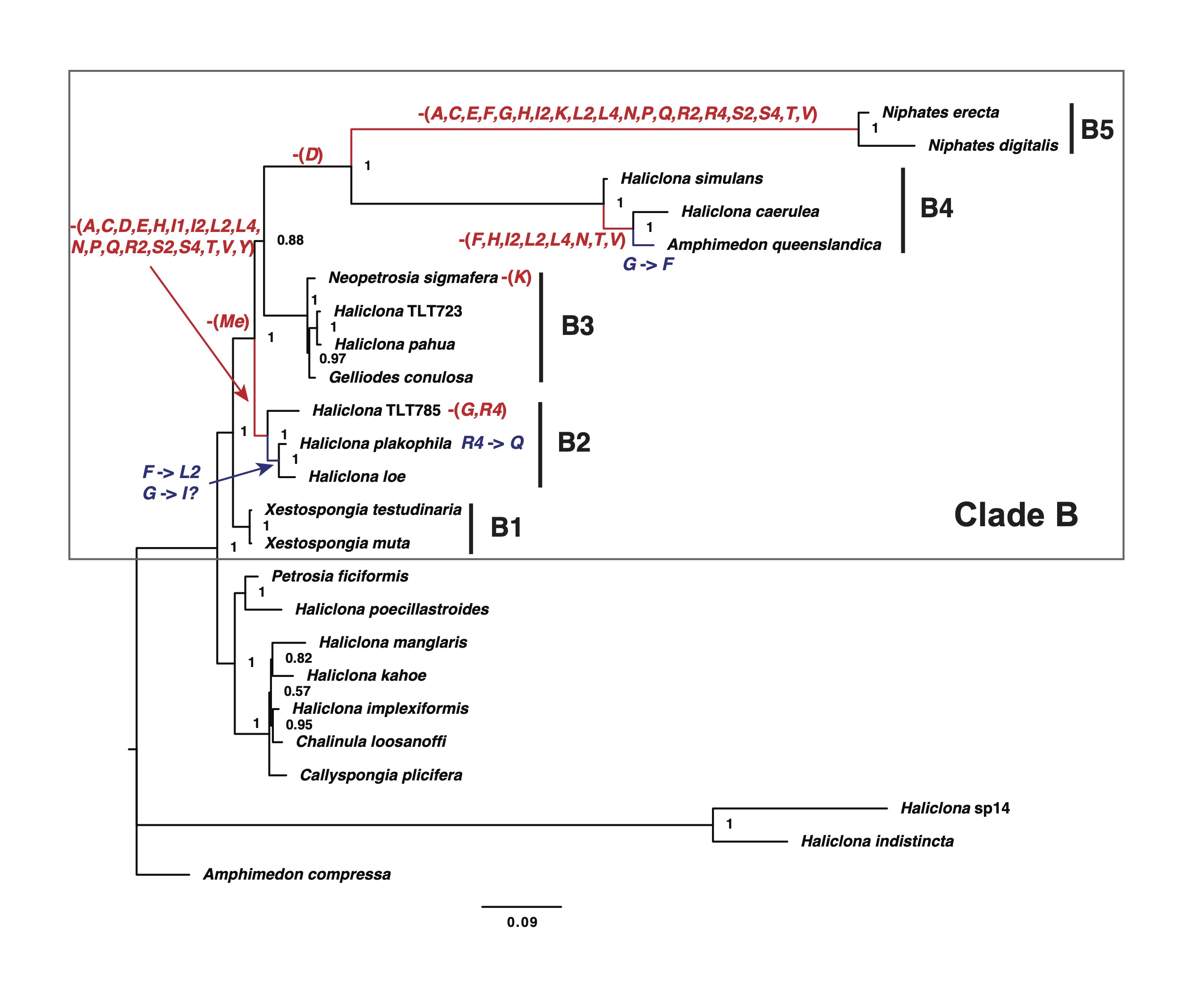
A group of sponges with a rampant mt-tRNA loss
Lavrov DV, Turner TL, and J Vicente
Genome Biology and Evolution, evaf020, 2025

Not entirely sure what a species is, but here are four new ones :)
Vicente J, Rutkowski E, Lavrov DV, Martineau G, Timmers M, and RJ Toonen
Zootaxa 5566(2), p243-272, 2025
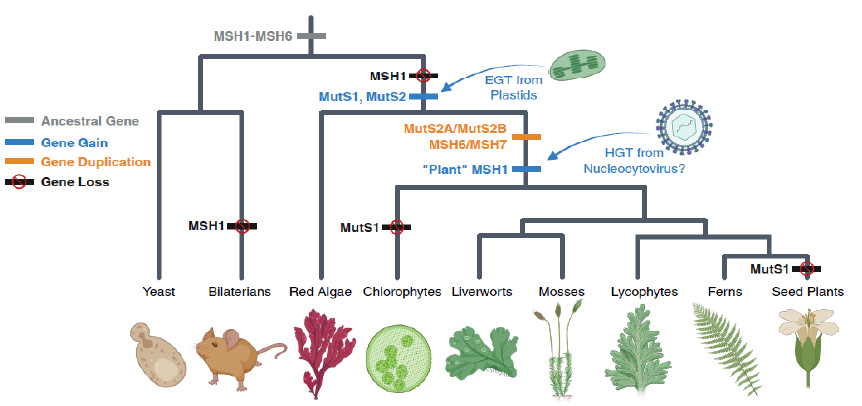
An adventure into the plant world!
Sloan DB, Broz AK, Kuster SA, Muthye V, Peñafiel-Ayala A, Marron JR, Lavrov DV, and LG Brieba
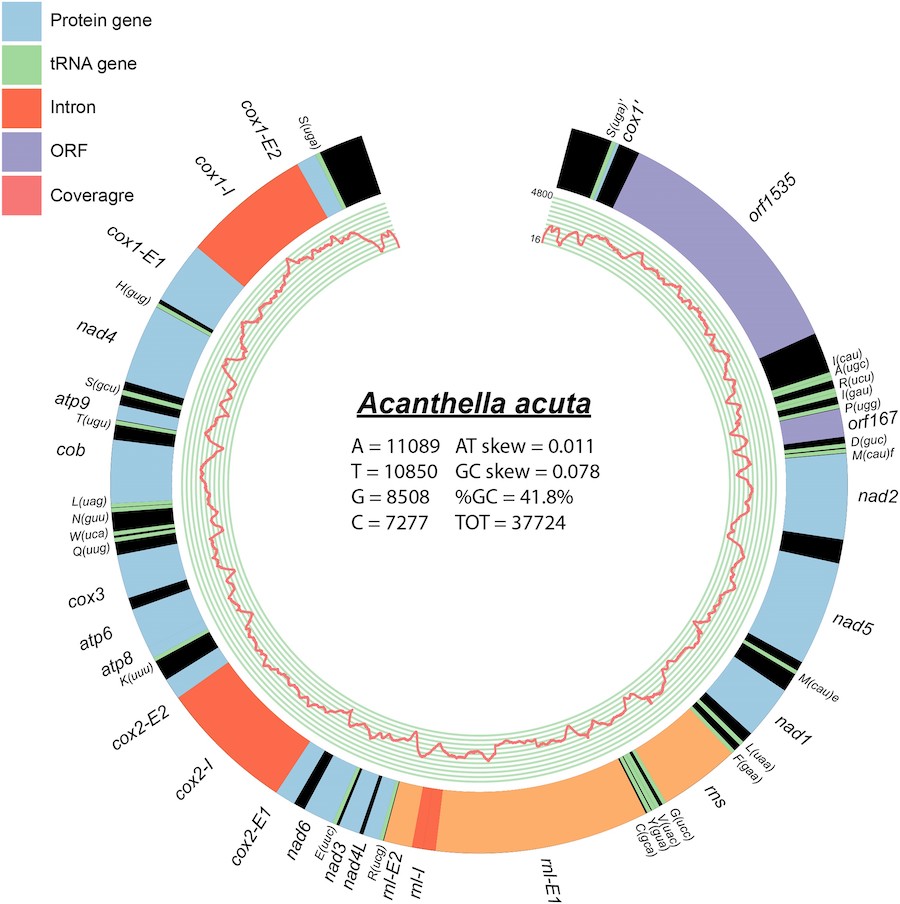
The fourth transition to a linear mtDNA architecture in animals
Ahmed M, Kayal E, and DV Lavrov
Genome Biology and Evolution, 16(8), 2024
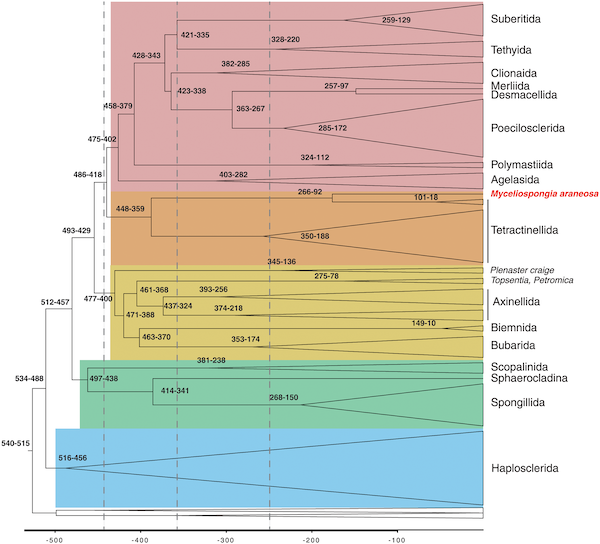
Demosponge phylogeny based on mtDNA data, including a very unexpected placement of Myceliospongia araneosa
Muthye V, Mackereth CD, Stewart JB, and DV Lavrov
PLoS One 18(12), e0287281, 2023
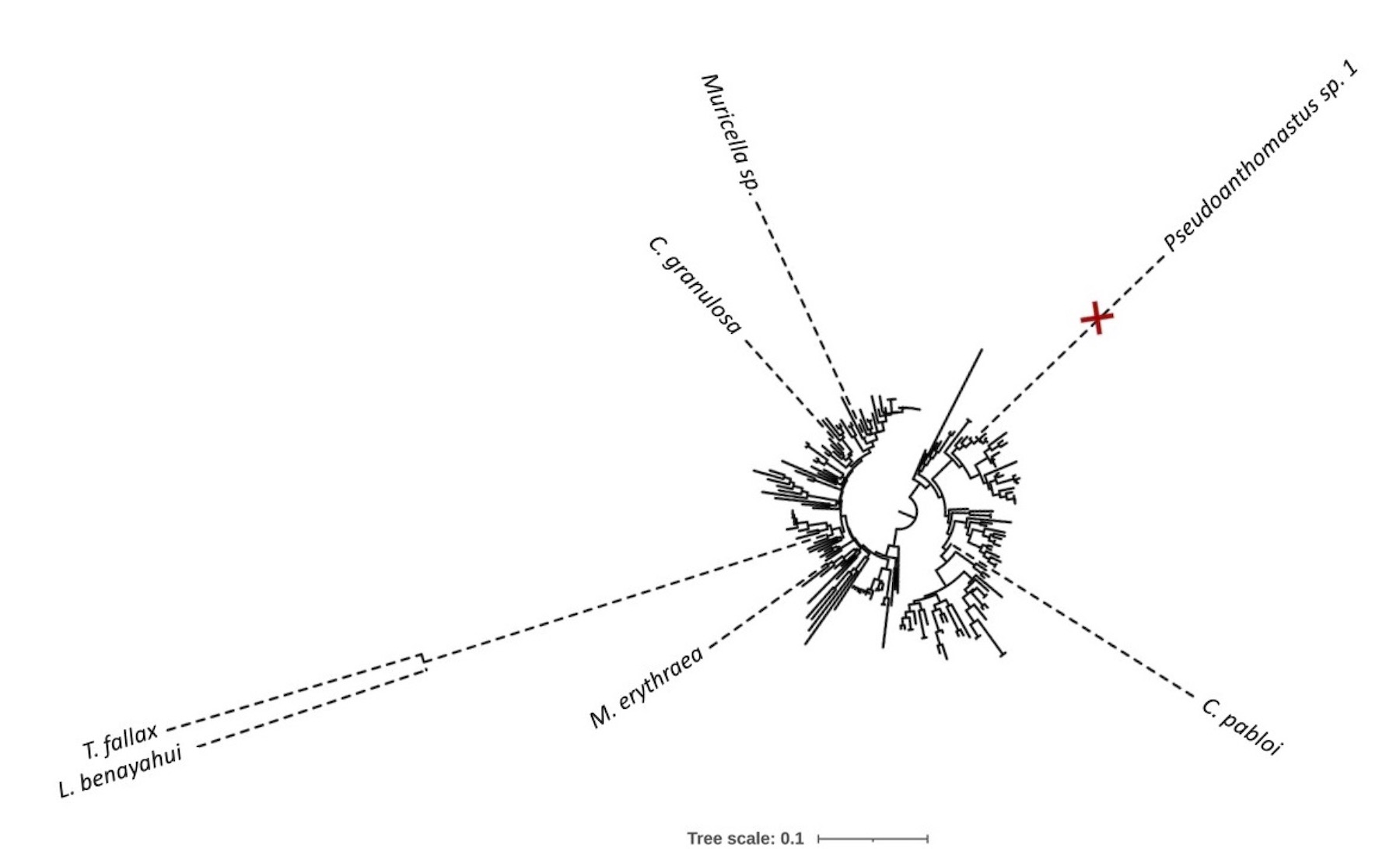
Among other things, the first octocoral mtDNA without mt-mutS
Muthye V, Mackereth CD, Stewart JB, and DV Lavrov
Recent
Pervasive mitochondrial tRNA gene loss in the clade B of haplosclerid sponges (Porifera, Demospongiae)
Lavrov DV, Turner TL, and J Vicente
Genome Biology and Evolution, evaf020, 2025
Integrative taxonomy of introduced Haplosclerida and four new species from Hawaii
Vicente J, Rutkowski E, Lavrov DV, Martineau G, Timmers M, and RJ Toonen
Zootaxa 5566(2), p243-272, 2025
Expansion of the MutS gene family in plants
Sloan DB, Broz AK, Kuster SA, Muthye V, Peñafiel-Ayala A, Marron JR, Lavrov DV, and LG Brieba
The Plant Cell, koae277, 2024
Mitochondrial DNA of the Demosponge Acanthella acuta: Linear Architecture and Other Unique Features
Ahmed M, Kayal E, and DV Lavrov
Genome Biology and Evolution, 16(8), 2024
Characterization of the Mitochondrial Proteome in the Ctenophore Mnemiopsis leidyi Using MitoPredictor
Muthye V, Mackereth CD, Stewart JB, and DV Lavrov
Ctenophores. Methods in Molecular Biology, vol 2757 pp 239-257, 2024
Phylomitogenomics bolsters the high-level classification of Demospongiae (phylum Porifera)
Muthye V, Mackereth CD, Stewart JB, and DV Lavrov
PLoS One 18(12), e0287281, 2023
Large dataset of octocoral mitochondrial genomes provides new insights into mt-mutS evolution and function
Muthye V, Mackereth CD, Stewart JB, and DV Lavrov
DNA repair, 110, 2022
Multiple losses of MSH1, gain of mtMutS, and other changes in the MutS family of DNA repair proteins in animals
Muthye V and DV Lavrov
Genome Biology and Evolution, 13 (9), 2021
Causes and consequences of mitochondrial proteome size variation in animals
Muthye V and DV Lavrov
Mitochondrion 52, 2020
MMPdb and MitoPredictor: Tools for facilitating comparative analysis of animal mitochondrial proteomes
Muthye V, Kandoi G, and DV Lavrov
Mitochondrion 51, 2020